pacman::p_load(sf, tidyverse, funModeling, blorr, corrplot, ggpubr, spdep, GWmodel, tmap, skimr, caret)In_class exercise 5
Geograpgically Weighted Logistic Regression (GWLR) and Application
Overview
In this lesson, you will learn the basic concepts and methods of logistic regression specially designed for geographical data. Upon completion of this lesson, you will able to:
explain the similarities and differences between Logistic Regression (LR) algorithm versus geographical weighted Logistic Regression (GWLR) algorithm.
calibrate predictive models by using appropriate Geographically Weighted Logistic Regression algorithm for geographical data.
Importing data
Importing water point data
First, we are going to import 2 set of data
wp <- readRDS("rds/osun_wp_sf.rds")glimpse(wp)Rows: 4,760
Columns: 75
$ row_id <dbl> 70578, 66401, 65607, 68989, 67708, 66419, 6…
$ source <chr> "Federal Ministry of Water Resources, Niger…
$ lat_deg <dbl> 7.759448, 8.031187, 7.891137, 7.509588, 7.4…
$ lon_deg <dbl> 4.563998, 4.637400, 4.713438, 4.265002, 4.3…
$ report_date <chr> "05/11/2015 12:00:00 AM", "04/30/2015 12:00…
$ status_id <chr> "No", "No", "No", "No", "Yes", "Yes", "Yes"…
$ water_source_clean <chr> "Borehole", "Borehole", "Borehole", "Boreho…
$ water_source_category <chr> "Well", "Well", "Well", "Well", "Well", "We…
$ water_tech_clean <chr> "Mechanized Pump", "Mechanized Pump", "Mech…
$ water_tech_category <chr> "Mechanized Pump", "Mechanized Pump", "Mech…
$ facility_type <chr> "Improved", "Improved", "Improved", "Improv…
$ clean_country_name <chr> "Nigeria", "Nigeria", "Nigeria", "Nigeria",…
$ clean_adm1 <chr> "Osun", "Osun", "Osun", "Osun", "Osun", "Os…
$ clean_adm2 <chr> "Osogbo", "Odo-Otin", "Boripe", "Ayedire", …
$ clean_adm3 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ clean_adm4 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ install_year <dbl> NA, 2004, 2006, 2014, 2011, 2007, 2007, 200…
$ installer <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ rehab_year <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ rehabilitator <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ management_clean <chr> NA, NA, NA, NA, NA, "Community Management",…
$ status_clean <chr> "Abandoned/Decommissioned", "Abandoned/Deco…
$ pay <chr> "No", "No", "No", "No", "No", "No", "No", "…
$ fecal_coliform_presence <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ fecal_coliform_value <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ subjective_quality <chr> "Acceptable quality", "Acceptable quality",…
$ activity_id <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ scheme_id <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ wpdx_id <chr> "6FV6QH57+QHH", "6FW62JJP+FXC", "6FV6VPR7+F…
$ notes <chr> "COSTAIN area, Osogbo", "Igbaye", "Araromi…
$ orig_lnk <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ photo_lnk <chr> "https://akvoflow-55.s3.amazonaws.com/image…
$ country_id <chr> "NG", "NG", "NG", "NG", "NG", "NG", "NG", "…
$ data_lnk <chr> "https://catalog.waterpointdata.org/dataset…
$ distance_to_primary_road <dbl> 167.82235, 4133.71306, 6559.76182, 8260.715…
$ distance_to_secondary_road <dbl> 838.9185, 1162.6246, 4625.0324, 5854.5731, …
$ distance_to_tertiary_road <dbl> 1181.107236, 9.012647, 58.314987, 2013.6515…
$ distance_to_city <dbl> 2449.2931, 16704.1923, 21516.8495, 31765.68…
$ distance_to_town <dbl> 9463.295, 5176.899, 8589.103, 16386.459, 23…
$ water_point_history <chr> "{\"2015-05-11\": {\"photo_lnk\": \"https:/…
$ rehab_priority <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ water_point_population <dbl> NA, NA, NA, NA, 0, 508, 162, 362, 3562, 217…
$ local_population_1km <dbl> NA, NA, NA, NA, 70, 647, 449, 1611, 7199, 2…
$ crucialness_score <dbl> NA, NA, NA, NA, NA, 0.785162287, 0.36080178…
$ pressure_score <dbl> NA, NA, NA, NA, NA, 1.69333333, 0.54000000,…
$ usage_capacity <dbl> 1000, 1000, 1000, 300, 1000, 300, 300, 300,…
$ is_urban <lgl> TRUE, FALSE, TRUE, FALSE, TRUE, FALSE, FALS…
$ days_since_report <dbl> 2689, 2700, 2688, 2693, 2701, 2692, 2681, 2…
$ staleness_score <dbl> 42.84384, 42.69554, 42.85735, 42.78986, 42.…
$ latest_record <lgl> TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, TRUE, T…
$ location_id <dbl> 239556, 230405, 240009, 236400, 229231, 237…
$ cluster_size <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 2…
$ clean_country_id <chr> "NGA", "NGA", "NGA", "NGA", "NGA", "NGA", "…
$ country_name <chr> "Nigeria", "Nigeria", "Nigeria", "Nigeria",…
$ water_source <chr> "Improved Tube well or borehole", "Improved…
$ water_tech <chr> "Motorised", "Motorised", "Motorised", "yet…
$ status <lgl> FALSE, FALSE, FALSE, FALSE, TRUE, TRUE, TRU…
$ adm2 <chr> "Osogbo", "Odo-Otin", "Boripe", "Aiyedire",…
$ adm3 <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ management <chr> NA, NA, NA, NA, NA, "Community Management",…
$ adm1 <chr> "Osun", "Osun", "Osun", "Osun", "Osun", "Os…
$ `New Georeferenced Column` <chr> "POINT (4.5639983 7.7594483)", "POINT (4.63…
$ lat_deg_original <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ lat_lon_deg <chr> "(7.7594483°, 4.5639983°)", "(8.0311867°, 4…
$ lon_deg_original <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA,…
$ public_data_source <chr> "https://catalog.waterpointdata.org/datafil…
$ converted <chr> "#status_id, #water_source, #pay, #status, …
$ count <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1…
$ created_timestamp <chr> "06/30/2020 12:56:07 PM", "06/30/2020 12:56…
$ updated_timestamp <chr> "06/30/2020 12:56:07 PM", "06/30/2020 12:56…
$ Geometry <POINT [m]> POINT (236239.7 417577), POINT (24463…
$ ADM2_EN <chr> "Osogbo", "Odo-Otin", "Boripe", "Aiyedire",…
$ ADM2_PCODE <chr> "NG030030", "NG030025", "NG030006", "NG0300…
$ ADM1_EN <chr> "Osun", "Osun", "Osun", "Osun", "Osun", "Os…
$ ADM1_PCODE <chr> "NG030", "NG030", "NG030", "NG030", "NG030"…osun <- readRDS("rds/Osun.rds")glimpse(osun)Rows: 30
Columns: 5
$ ADM2_EN <chr> "Aiyedade", "Aiyedire", "Atakumosa East", "Atakumosa West",…
$ ADM2_PCODE <chr> "NG030001", "NG030002", "NG030003", "NG030004", "NG030005",…
$ ADM1_EN <chr> "Osun", "Osun", "Osun", "Osun", "Osun", "Osun", "Osun", "Os…
$ ADM1_PCODE <chr> "NG030", "NG030", "NG030", "NG030", "NG030", "NG030", "NG03…
$ geometry <MULTIPOLYGON [m]> MULTIPOLYGON (((213526.6 34..., MULTIPOLYGON (…tm_shape(osun) +
tm_polygons()+
tm_shape(wp)+
tm_dots()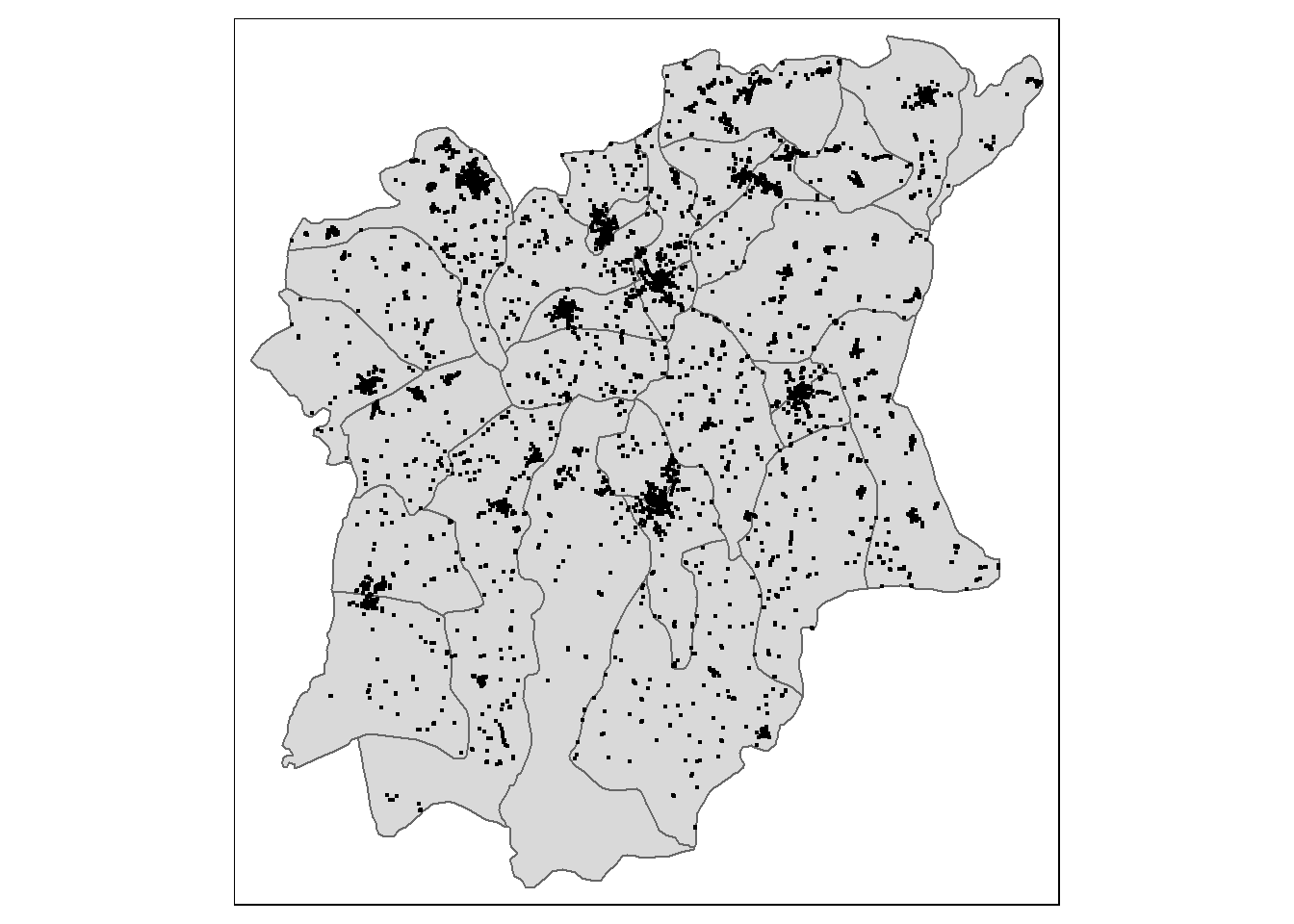
Exploratory Data Analysis
freq(wp, input='status')Warning: The `<scale>` argument of `guides()` cannot be `FALSE`. Use "none" instead as
of ggplot2 3.3.4.
ℹ The deprecated feature was likely used in the funModeling package.
Please report the issue at <https://github.com/pablo14/funModeling/issues>.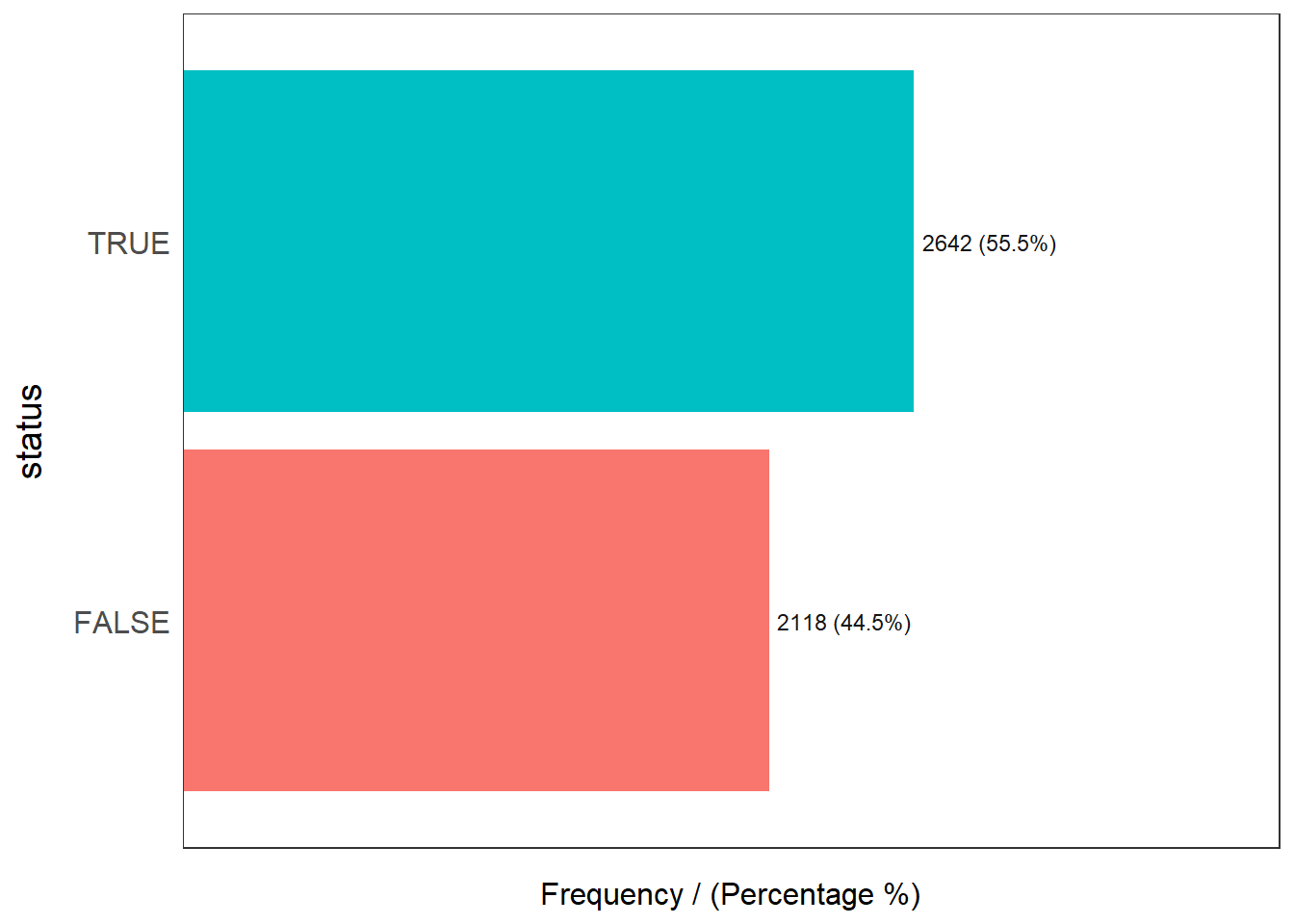
status frequency percentage cumulative_perc
1 TRUE 2642 55.5 55.5
2 FALSE 2118 44.5 100.0tmap_mode("view")tmap mode set to interactive viewingtmap mode set to interactive viewing
tm_shape(osun) +
tm_polygons(alpha=0.4) +
tm_shape(wp) +
tm_dots(col="status",
alpha=0.6) +
tm_view(set.zoom.limits = c(9, 12))the skim() function of the skimr package to do quick exploratory data analysis. For categorical variables, it shows the number of the missing values and unique variable view. For binary variables, shows the number of missing values and gives a frequency count. For numercial fields, on top of missing values, it also shows some summary statistics like mean, standard deviation.
skim(wp)Warning: Couldn't find skimmers for class: sfc_POINT, sfc; No user-defined `sfl`
provided. Falling back to `character`.| Name | wp |
| Number of rows | 4760 |
| Number of columns | 75 |
| _______________________ | |
| Column type frequency: | |
| character | 47 |
| logical | 5 |
| numeric | 23 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| source | 0 | 1.00 | 5 | 44 | 0 | 2 | 0 |
| report_date | 0 | 1.00 | 22 | 22 | 0 | 42 | 0 |
| status_id | 0 | 1.00 | 2 | 7 | 0 | 3 | 0 |
| water_source_clean | 0 | 1.00 | 8 | 22 | 0 | 3 | 0 |
| water_source_category | 0 | 1.00 | 4 | 6 | 0 | 2 | 0 |
| water_tech_clean | 24 | 0.99 | 9 | 23 | 0 | 3 | 0 |
| water_tech_category | 24 | 0.99 | 9 | 15 | 0 | 2 | 0 |
| facility_type | 0 | 1.00 | 8 | 8 | 0 | 1 | 0 |
| clean_country_name | 0 | 1.00 | 7 | 7 | 0 | 1 | 0 |
| clean_adm1 | 0 | 1.00 | 3 | 5 | 0 | 5 | 0 |
| clean_adm2 | 0 | 1.00 | 3 | 14 | 0 | 35 | 0 |
| clean_adm3 | 4760 | 0.00 | NA | NA | 0 | 0 | 0 |
| clean_adm4 | 4760 | 0.00 | NA | NA | 0 | 0 | 0 |
| installer | 4760 | 0.00 | NA | NA | 0 | 0 | 0 |
| management_clean | 1573 | 0.67 | 5 | 37 | 0 | 7 | 0 |
| status_clean | 0 | 1.00 | 9 | 32 | 0 | 7 | 0 |
| pay | 0 | 1.00 | 2 | 39 | 0 | 7 | 0 |
| fecal_coliform_presence | 4760 | 0.00 | NA | NA | 0 | 0 | 0 |
| subjective_quality | 0 | 1.00 | 18 | 20 | 0 | 4 | 0 |
| activity_id | 4757 | 0.00 | 36 | 36 | 0 | 3 | 0 |
| scheme_id | 4760 | 0.00 | NA | NA | 0 | 0 | 0 |
| wpdx_id | 0 | 1.00 | 12 | 12 | 0 | 4760 | 0 |
| notes | 0 | 1.00 | 2 | 96 | 0 | 3502 | 0 |
| orig_lnk | 4757 | 0.00 | 84 | 84 | 0 | 1 | 0 |
| photo_lnk | 41 | 0.99 | 84 | 84 | 0 | 4719 | 0 |
| country_id | 0 | 1.00 | 2 | 2 | 0 | 1 | 0 |
| data_lnk | 0 | 1.00 | 79 | 96 | 0 | 2 | 0 |
| water_point_history | 0 | 1.00 | 142 | 834 | 0 | 4750 | 0 |
| clean_country_id | 0 | 1.00 | 3 | 3 | 0 | 1 | 0 |
| country_name | 0 | 1.00 | 7 | 7 | 0 | 1 | 0 |
| water_source | 0 | 1.00 | 8 | 30 | 0 | 4 | 0 |
| water_tech | 0 | 1.00 | 5 | 37 | 0 | 20 | 0 |
| adm2 | 0 | 1.00 | 3 | 14 | 0 | 33 | 0 |
| adm3 | 4760 | 0.00 | NA | NA | 0 | 0 | 0 |
| management | 1573 | 0.67 | 5 | 47 | 0 | 7 | 0 |
| adm1 | 0 | 1.00 | 4 | 5 | 0 | 4 | 0 |
| New Georeferenced Column | 0 | 1.00 | 16 | 35 | 0 | 4760 | 0 |
| lat_lon_deg | 0 | 1.00 | 13 | 32 | 0 | 4760 | 0 |
| public_data_source | 0 | 1.00 | 84 | 102 | 0 | 2 | 0 |
| converted | 0 | 1.00 | 53 | 53 | 0 | 1 | 0 |
| created_timestamp | 0 | 1.00 | 22 | 22 | 0 | 2 | 0 |
| updated_timestamp | 0 | 1.00 | 22 | 22 | 0 | 2 | 0 |
| Geometry | 0 | 1.00 | 33 | 37 | 0 | 4760 | 0 |
| ADM2_EN | 0 | 1.00 | 3 | 14 | 0 | 30 | 0 |
| ADM2_PCODE | 0 | 1.00 | 8 | 8 | 0 | 30 | 0 |
| ADM1_EN | 0 | 1.00 | 4 | 4 | 0 | 1 | 0 |
| ADM1_PCODE | 0 | 1.00 | 5 | 5 | 0 | 1 | 0 |
Variable type: logical
| skim_variable | n_missing | complete_rate | mean | count |
|---|---|---|---|---|
| rehab_year | 4760 | 0 | NaN | : |
| rehabilitator | 4760 | 0 | NaN | : |
| is_urban | 0 | 1 | 0.39 | FAL: 2884, TRU: 1876 |
| latest_record | 0 | 1 | 1.00 | TRU: 4760 |
| status | 0 | 1 | 0.56 | TRU: 2642, FAL: 2118 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| row_id | 0 | 1.00 | 68550.48 | 10216.94 | 49601.00 | 66874.75 | 68244.50 | 69562.25 | 471319.00 | ▇▁▁▁▁ |
| lat_deg | 0 | 1.00 | 7.68 | 0.22 | 7.06 | 7.51 | 7.71 | 7.88 | 8.06 | ▁▂▇▇▇ |
| lon_deg | 0 | 1.00 | 4.54 | 0.21 | 4.08 | 4.36 | 4.56 | 4.71 | 5.06 | ▃▆▇▇▂ |
| install_year | 1144 | 0.76 | 2008.63 | 6.04 | 1917.00 | 2006.00 | 2010.00 | 2013.00 | 2015.00 | ▁▁▁▁▇ |
| fecal_coliform_value | 4760 | 0.00 | NaN | NA | NA | NA | NA | NA | NA | |
| distance_to_primary_road | 0 | 1.00 | 5021.53 | 5648.34 | 0.01 | 719.36 | 2972.78 | 7314.73 | 26909.86 | ▇▂▁▁▁ |
| distance_to_secondary_road | 0 | 1.00 | 3750.47 | 3938.63 | 0.15 | 460.90 | 2554.25 | 5791.94 | 19559.48 | ▇▃▁▁▁ |
| distance_to_tertiary_road | 0 | 1.00 | 1259.28 | 1680.04 | 0.02 | 121.25 | 521.77 | 1834.42 | 10966.27 | ▇▂▁▁▁ |
| distance_to_city | 0 | 1.00 | 16663.99 | 10960.82 | 53.05 | 7930.75 | 15030.41 | 24255.75 | 47934.34 | ▇▇▆▃▁ |
| distance_to_town | 0 | 1.00 | 16726.59 | 12452.65 | 30.00 | 6876.92 | 12204.53 | 27739.46 | 44020.64 | ▇▅▃▃▂ |
| rehab_priority | 2654 | 0.44 | 489.33 | 1658.81 | 0.00 | 7.00 | 91.50 | 376.25 | 29697.00 | ▇▁▁▁▁ |
| water_point_population | 4 | 1.00 | 513.58 | 1458.92 | 0.00 | 14.00 | 119.00 | 433.25 | 29697.00 | ▇▁▁▁▁ |
| local_population_1km | 4 | 1.00 | 2727.16 | 4189.46 | 0.00 | 176.00 | 1032.00 | 3717.00 | 36118.00 | ▇▁▁▁▁ |
| crucialness_score | 798 | 0.83 | 0.26 | 0.28 | 0.00 | 0.07 | 0.15 | 0.35 | 1.00 | ▇▃▁▁▁ |
| pressure_score | 798 | 0.83 | 1.46 | 4.16 | 0.00 | 0.12 | 0.41 | 1.24 | 93.69 | ▇▁▁▁▁ |
| usage_capacity | 0 | 1.00 | 560.74 | 338.46 | 300.00 | 300.00 | 300.00 | 1000.00 | 1000.00 | ▇▁▁▁▅ |
| days_since_report | 0 | 1.00 | 2692.69 | 41.92 | 1483.00 | 2688.00 | 2693.00 | 2700.00 | 4645.00 | ▁▇▁▁▁ |
| staleness_score | 0 | 1.00 | 42.80 | 0.58 | 23.13 | 42.70 | 42.79 | 42.86 | 62.66 | ▁▁▇▁▁ |
| location_id | 0 | 1.00 | 235865.49 | 6657.60 | 23741.00 | 230638.75 | 236199.50 | 240061.25 | 267454.00 | ▁▁▁▁▇ |
| cluster_size | 0 | 1.00 | 1.05 | 0.25 | 1.00 | 1.00 | 1.00 | 1.00 | 4.00 | ▇▁▁▁▁ |
| lat_deg_original | 4760 | 0.00 | NaN | NA | NA | NA | NA | NA | NA | |
| lon_deg_original | 4760 | 0.00 | NaN | NA | NA | NA | NA | NA | NA | |
| count | 0 | 1.00 | 1.00 | 0.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | ▁▁▇▁▁ |
expvars <- c("status","distance_to_primary_road", "distance_to_secondary_road",
"distance_to_tertiary_road", "distance_to_city",
"distance_to_town", "water_point_population",
"local_population_1km", "usage_capacity", "is_urban",
"water_source_clean")
wp_clean <- wp %>%
filter(!if_any(expvars, ~is.na(.x)))%>%
mutate(usage_capacity = as.factor(usage_capacity))Warning: Using an external vector in selections was deprecated in tidyselect 1.1.0.
ℹ Please use `all_of()` or `any_of()` instead.
# Was:
data %>% select(expvars)
# Now:
data %>% select(all_of(expvars))
See <https://tidyselect.r-lib.org/reference/faq-external-vector.html>.Next, we need to check for multicollinearity. As we cannot o that on the data with geometry information, we need to strip the geometry information with st_set_geometry(NULL). We can then plot the correlation matrix.
wp_vars <- wp_clean %>%
select(expvars)%>%
st_set_geometry(NULL)vars.cor = cor(wp_vars[2:7])
corrplot.mixed(vars.cor,
lower="ellipse",
upper="number",
tl.pos = "lt",
diag="l",
tl.col = "black")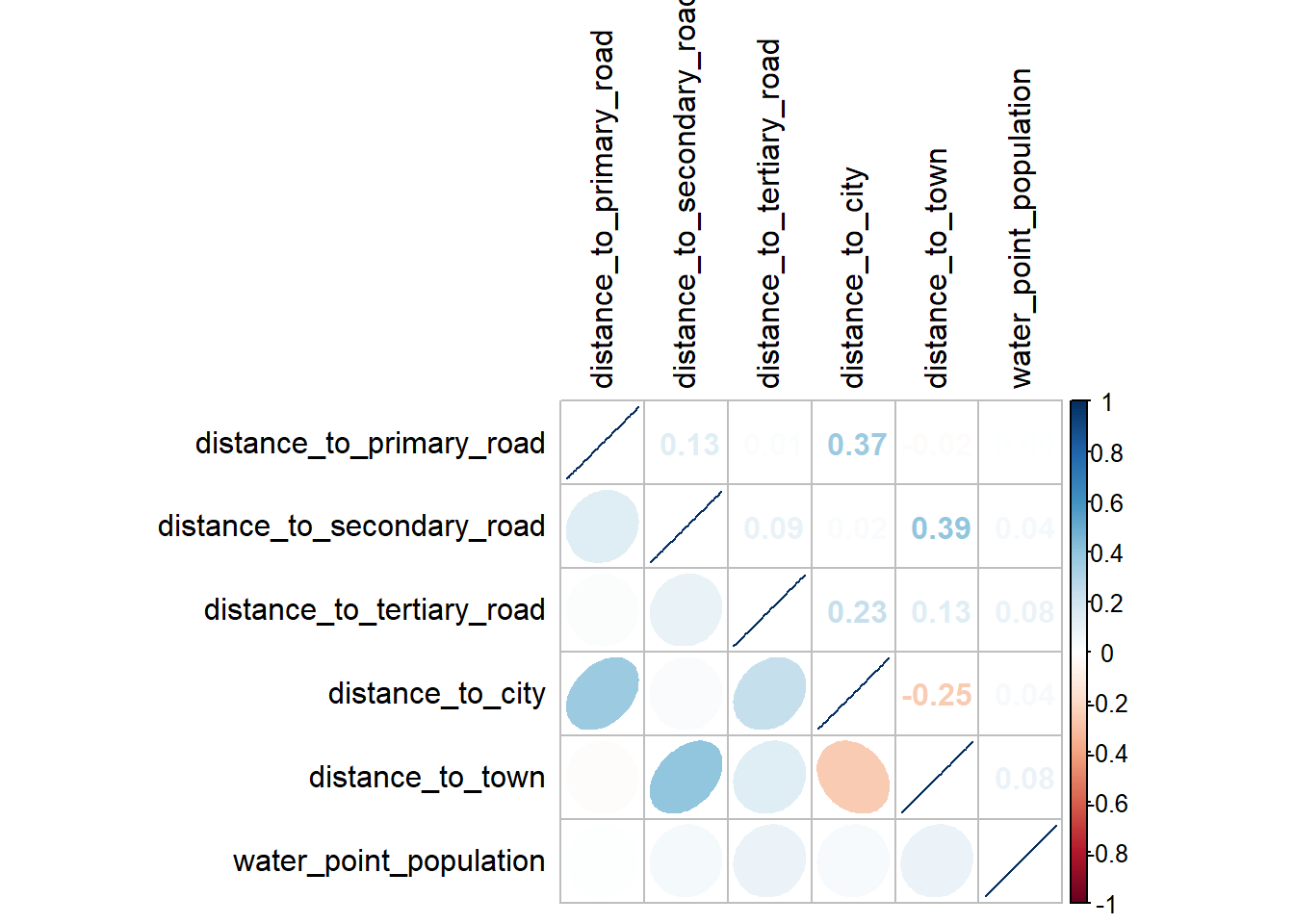
There are no variables that display multi-collinearity so we do not need to drop any. Now we can proceed to do a logistic regression. The first line of the code below creates the regression formula from the list of variables of interest using the paste() function. This method of creating a formula from lists is convenient when we have many explanatory variables so we don’t need to keep typing it out. We can then input the formula into the glm() function to perform the regression.
fm <- as.formula(paste("status ~",
paste(expvars[2:11], collapse="+")))
model <- glm(fm,
data=wp_clean,
family=binomial(link="logit"))The blr_regress() function of the blorr package creates a neat logistic regression report.
blr_regress(model) Model Overview
------------------------------------------------------------------------
Data Set Resp Var Obs. Df. Model Df. Residual Convergence
------------------------------------------------------------------------
data status 4756 4755 4744 TRUE
------------------------------------------------------------------------
Response Summary
--------------------------------------------------------
Outcome Frequency Outcome Frequency
--------------------------------------------------------
0 2114 1 2642
--------------------------------------------------------
Maximum Likelihood Estimates
-----------------------------------------------------------------------------------------------
Parameter DF Estimate Std. Error z value Pr(>|z|)
-----------------------------------------------------------------------------------------------
(Intercept) 1 0.3887 0.1124 3.4588 5e-04
distance_to_primary_road 1 0.0000 0.0000 -0.7153 0.4744
distance_to_secondary_road 1 0.0000 0.0000 -0.5530 0.5802
distance_to_tertiary_road 1 1e-04 0.0000 4.6708 0.0000
distance_to_city 1 0.0000 0.0000 -4.7574 0.0000
distance_to_town 1 0.0000 0.0000 -4.9170 0.0000
water_point_population 1 -5e-04 0.0000 -11.3686 0.0000
local_population_1km 1 3e-04 0.0000 19.2953 0.0000
usage_capacity1000 1 -0.6230 0.0697 -8.9366 0.0000
is_urbanTRUE 1 -0.2971 0.0819 -3.6294 3e-04
water_source_cleanProtected Shallow Well 1 0.5040 0.0857 5.8783 0.0000
water_source_cleanProtected Spring 1 1.2882 0.4388 2.9359 0.0033
-----------------------------------------------------------------------------------------------
Association of Predicted Probabilities and Observed Responses
---------------------------------------------------------------
% Concordant 0.7347 Somers' D 0.4693
% Discordant 0.2653 Gamma 0.4693
% Tied 0.0000 Tau-a 0.2318
Pairs 5585188 c 0.7347
---------------------------------------------------------------There are 2 variables which are not statistically significant (p-value>0.05). They are not good predictors and should be considered. distance_to_tertiary_road, distance_to_city, distance_to_town and local_population_1km have positive coefficients, indicating that larger values correspond with higher possibility of a waterpoint being functional.
blr_confusion_matrix(model, cutoff=0.5)Confusion Matrix and Statistics
Reference
Prediction FALSE TRUE
0 1301 738
1 813 1904
Accuracy : 0.6739
No Information Rate : 0.4445
Kappa : 0.3373
McNemars's Test P-Value : 0.0602
Sensitivity : 0.7207
Specificity : 0.6154
Pos Pred Value : 0.7008
Neg Pred Value : 0.6381
Prevalence : 0.5555
Detection Rate : 0.4003
Detection Prevalence : 0.5713
Balanced Accuracy : 0.6680
Precision : 0.7008
Recall : 0.7207
'Positive' Class : 1The overall accuracy of the model is 67%. The model is better at predicting positives than negatives as the true positive rate (sensitivity) is higher than the true negative rate (specificity).
Geographically Weighted Logistic Regression
wp_clean_sp <- wp_clean %>%
select(expvars) %>%
as_Spatial()
wp_clean_spclass : SpatialPointsDataFrame
features : 4756
extent : 182502.4, 290751, 340054.1, 450905.3 (xmin, xmax, ymin, ymax)
crs : +proj=tmerc +lat_0=4 +lon_0=8.5 +k=0.99975 +x_0=670553.98 +y_0=0 +a=6378249.145 +rf=293.465 +towgs84=-92,-93,122,0,0,0,0 +units=m +no_defs
variables : 11
names : status, distance_to_primary_road, distance_to_secondary_road, distance_to_tertiary_road, distance_to_city, distance_to_town, water_point_population, local_population_1km, usage_capacity, is_urban, water_source_clean
min values : 0, 0.014461356813335, 0.152195902540837, 0.017815121653488, 53.0461399623541, 30.0019777713073, 0, 0, 1000, 0, Borehole
max values : 1, 26909.8616132094, 19559.4793799085, 10966.2705628969, 47934.343603562, 44020.6393368124, 29697, 36118, 300, 1, Protected Spring The next step is to create the spatial weights matrix. We need to use a distance-based spatial weights matrix to conduct the logistic regression. The following code chunk uses AIC to recommend the maximum distance to consider neighbours for a fixed distance matrix.
bw.fixed <- bw.ggwr(fm,
data=wp_clean_sp,
family = "binomial",
approach= "AIC",
kernel = "gaussian",
adaptive = FALSE,
longlat= FALSE)Take a cup of tea and have a break, it will take a few minutes.
-----A kind suggestion from GWmodel development group
Iteration Log-Likelihood:(With bandwidth: 95768.67 )
=========================
0 -2889
1 -2836
2 -2830
3 -2829
4 -2829
5 -2829
Fixed bandwidth: 95768.67 AICc value: 5684.357
Iteration Log-Likelihood:(With bandwidth: 59200.13 )
=========================
0 -2875
1 -2818
2 -2810
3 -2808
4 -2808
5 -2808
Fixed bandwidth: 59200.13 AICc value: 5646.785
Iteration Log-Likelihood:(With bandwidth: 36599.53 )
=========================
0 -2847
1 -2781
2 -2768
3 -2765
4 -2765
5 -2765
6 -2765
Fixed bandwidth: 36599.53 AICc value: 5575.148
Iteration Log-Likelihood:(With bandwidth: 22631.59 )
=========================
0 -2798
1 -2719
2 -2698
3 -2693
4 -2693
5 -2693
6 -2693
Fixed bandwidth: 22631.59 AICc value: 5466.883
Iteration Log-Likelihood:(With bandwidth: 13998.93 )
=========================
0 -2720
1 -2622
2 -2590
3 -2581
4 -2580
5 -2580
6 -2580
7 -2580
Fixed bandwidth: 13998.93 AICc value: 5324.578
Iteration Log-Likelihood:(With bandwidth: 8663.649 )
=========================
0 -2601
1 -2476
2 -2431
3 -2419
4 -2417
5 -2417
6 -2417
7 -2417
Fixed bandwidth: 8663.649 AICc value: 5163.61
Iteration Log-Likelihood:(With bandwidth: 5366.266 )
=========================
0 -2436
1 -2268
2 -2194
3 -2167
4 -2161
5 -2161
6 -2161
7 -2161
8 -2161
9 -2161
Fixed bandwidth: 5366.266 AICc value: 4990.587
Iteration Log-Likelihood:(With bandwidth: 3328.371 )
=========================
0 -2157
1 -1922
2 -1802
3 -1739
4 -1713
5 -1713
Fixed bandwidth: 3328.371 AICc value: 4798.288
Iteration Log-Likelihood:(With bandwidth: 2068.882 )
=========================
0 -1751
1 -1421
2 -1238
3 -1133
4 -1084
5 -1084
Fixed bandwidth: 2068.882 AICc value: 4837.017
Iteration Log-Likelihood:(With bandwidth: 4106.777 )
=========================
0 -2297
1 -2095
2 -1997
3 -1951
4 -1938
5 -1936
6 -1936
7 -1936
8 -1936
Fixed bandwidth: 4106.777 AICc value: 4873.161
Iteration Log-Likelihood:(With bandwidth: 2847.289 )
=========================
0 -2036
1 -1771
2 -1633
3 -1558
4 -1525
5 -1525
Fixed bandwidth: 2847.289 AICc value: 4768.192
Iteration Log-Likelihood:(With bandwidth: 2549.964 )
=========================
0 -1941
1 -1655
2 -1503
3 -1417
4 -1378
5 -1378
Fixed bandwidth: 2549.964 AICc value: 4762.212
Iteration Log-Likelihood:(With bandwidth: 2366.207 )
=========================
0 -1874
1 -1573
2 -1410
3 -1316
4 -1274
5 -1274
Fixed bandwidth: 2366.207 AICc value: 4773.081
Iteration Log-Likelihood:(With bandwidth: 2663.532 )
=========================
0 -1979
1 -1702
2 -1555
3 -1474
4 -1438
5 -1438
Fixed bandwidth: 2663.532 AICc value: 4762.568
Iteration Log-Likelihood:(With bandwidth: 2479.775 )
=========================
0 -1917
1 -1625
2 -1468
3 -1380
4 -1339
5 -1339
Fixed bandwidth: 2479.775 AICc value: 4764.294
Iteration Log-Likelihood:(With bandwidth: 2593.343 )
=========================
0 -1956
1 -1674
2 -1523
3 -1439
4 -1401
5 -1401
Fixed bandwidth: 2593.343 AICc value: 4761.813
Iteration Log-Likelihood:(With bandwidth: 2620.153 )
=========================
0 -1965
1 -1685
2 -1536
3 -1453
4 -1415
5 -1415
Fixed bandwidth: 2620.153 AICc value: 4761.89
Iteration Log-Likelihood:(With bandwidth: 2576.774 )
=========================
0 -1950
1 -1667
2 -1515
3 -1431
4 -1393
5 -1393
Fixed bandwidth: 2576.774 AICc value: 4761.889
Iteration Log-Likelihood:(With bandwidth: 2603.584 )
=========================
0 -1960
1 -1678
2 -1528
3 -1445
4 -1407
5 -1407
Fixed bandwidth: 2603.584 AICc value: 4761.813
Iteration Log-Likelihood:(With bandwidth: 2609.913 )
=========================
0 -1962
1 -1680
2 -1531
3 -1448
4 -1410
5 -1410
Fixed bandwidth: 2609.913 AICc value: 4761.831
Iteration Log-Likelihood:(With bandwidth: 2599.672 )
=========================
0 -1958
1 -1676
2 -1526
3 -1443
4 -1405
5 -1405
Fixed bandwidth: 2599.672 AICc value: 4761.809
Iteration Log-Likelihood:(With bandwidth: 2597.255 )
=========================
0 -1957
1 -1675
2 -1525
3 -1441
4 -1403
5 -1403
Fixed bandwidth: 2597.255 AICc value: 4761.809 bw.fixed[1] 2599.672The recommended maximum bandwidth for fixed distance matrix is 2599.672m.
gwlr.fixed <- ggwr.basic(fm,
data=wp_clean_sp,
bw = 2599.672,
family = "binomial",
kernel = "gaussian",
adaptive = FALSE,
longlat= FALSE) Iteration Log-Likelihood
=========================
0 -1958
1 -1676
2 -1526
3 -1443
4 -1405
5 -1405 gwlr.fixed ***********************************************************************
* Package GWmodel *
***********************************************************************
Program starts at: 2022-12-18 12:28:12
Call:
ggwr.basic(formula = fm, data = wp_clean_sp, bw = 2599.672, family = "binomial",
kernel = "gaussian", adaptive = FALSE, longlat = FALSE)
Dependent (y) variable: status
Independent variables: distance_to_primary_road distance_to_secondary_road distance_to_tertiary_road distance_to_city distance_to_town water_point_population local_population_1km usage_capacity is_urban water_source_clean
Number of data points: 4756
Used family: binomial
***********************************************************************
* Results of Generalized linear Regression *
***********************************************************************
Call:
NULL
Deviance Residuals:
Min 1Q Median 3Q Max
-124.555 -1.755 1.072 1.742 34.333
Coefficients:
Estimate Std. Error z value Pr(>|z|)
Intercept 3.887e-01 1.124e-01 3.459 0.000543
distance_to_primary_road -4.642e-06 6.490e-06 -0.715 0.474422
distance_to_secondary_road -5.143e-06 9.299e-06 -0.553 0.580230
distance_to_tertiary_road 9.683e-05 2.073e-05 4.671 3.00e-06
distance_to_city -1.686e-05 3.544e-06 -4.757 1.96e-06
distance_to_town -1.480e-05 3.009e-06 -4.917 8.79e-07
water_point_population -5.097e-04 4.484e-05 -11.369 < 2e-16
local_population_1km 3.451e-04 1.788e-05 19.295 < 2e-16
usage_capacity1000 -6.230e-01 6.972e-02 -8.937 < 2e-16
is_urbanTRUE -2.971e-01 8.185e-02 -3.629 0.000284
water_source_cleanProtected Shallow Well 5.040e-01 8.574e-02 5.878 4.14e-09
water_source_cleanProtected Spring 1.288e+00 4.388e-01 2.936 0.003325
Intercept ***
distance_to_primary_road
distance_to_secondary_road
distance_to_tertiary_road ***
distance_to_city ***
distance_to_town ***
water_point_population ***
local_population_1km ***
usage_capacity1000 ***
is_urbanTRUE ***
water_source_cleanProtected Shallow Well ***
water_source_cleanProtected Spring **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 6534.5 on 4755 degrees of freedom
Residual deviance: 5688.0 on 4744 degrees of freedom
AIC: 5712
Number of Fisher Scoring iterations: 5
AICc: 5712.099
Pseudo R-square value: 0.1295351
***********************************************************************
* Results of Geographically Weighted Regression *
***********************************************************************
*********************Model calibration information*********************
Kernel function: gaussian
Fixed bandwidth: 2599.672
Regression points: the same locations as observations are used.
Distance metric: A distance matrix is specified for this model calibration.
************Summary of Generalized GWR coefficient estimates:**********
Min. 1st Qu. Median
Intercept -8.7229e+02 -4.9955e+00 1.7600e+00
distance_to_primary_road -1.9389e-02 -4.8031e-04 2.9618e-05
distance_to_secondary_road -1.5921e-02 -3.7551e-04 1.2317e-04
distance_to_tertiary_road -1.5618e-02 -4.2368e-04 7.6179e-05
distance_to_city -1.8416e-02 -5.6217e-04 -1.2726e-04
distance_to_town -2.2411e-02 -5.7283e-04 -1.5155e-04
water_point_population -5.2208e-02 -2.2767e-03 -9.8875e-04
local_population_1km -1.2698e-01 4.9952e-04 1.0638e-03
usage_capacity1000 -2.0772e+01 -9.7231e-01 -4.1592e-01
is_urbanTRUE -1.9790e+02 -4.2908e+00 -1.6864e+00
water_source_cleanProtected.Shallow.Well -2.0789e+01 -4.5190e-01 5.3340e-01
water_source_cleanProtected.Spring -5.2235e+02 -5.5977e+00 2.5441e+00
3rd Qu. Max.
Intercept 1.2763e+01 1073.2156
distance_to_primary_road 4.8443e-04 0.0142
distance_to_secondary_road 6.0692e-04 0.0258
distance_to_tertiary_road 6.6815e-04 0.0128
distance_to_city 2.3718e-04 0.0150
distance_to_town 1.9271e-04 0.0224
water_point_population 5.0102e-04 0.1309
local_population_1km 1.8157e-03 0.0392
usage_capacity1000 3.0322e-01 5.9281
is_urbanTRUE 1.2841e+00 744.3099
water_source_cleanProtected.Shallow.Well 1.7849e+00 67.6343
water_source_cleanProtected.Spring 6.7663e+00 317.4133
************************Diagnostic information*************************
Number of data points: 4756
GW Deviance: 2795.084
AIC : 4414.606
AICc : 4747.423
Pseudo R-square value: 0.5722559
***********************************************************************
Program stops at: 2022-12-18 12:32:20 In order to assess the performance of the GWLR, we need to extract the output into a dataframe.
gwr.fixed <- as.data.frame(gwlr.fixed$SDF)We manually compute the predicted waterpoint status from the probability that the waterpoint is functional (yhat) using the threshold of 0.5 again.
gwr.fixed <- gwr.fixed %>%
mutate(most = ifelse(
yhat >= 0.5, T, F
))The following code chunk creates a confusion matrix by comparing the actual outcome with the predicted likely outcome.
gwr.fixed$y <- as.factor(gwr.fixed$y)
gwr.fixed$most <- as.factor(gwr.fixed$most)
CM <- confusionMatrix(data=gwr.fixed$most,
positive= "TRUE",
reference = gwr.fixed$y)
CMConfusion Matrix and Statistics
Reference
Prediction FALSE TRUE
FALSE 1824 263
TRUE 290 2379
Accuracy : 0.8837
95% CI : (0.8743, 0.8927)
No Information Rate : 0.5555
P-Value [Acc > NIR] : <2e-16
Kappa : 0.7642
Mcnemar's Test P-Value : 0.2689
Sensitivity : 0.9005
Specificity : 0.8628
Pos Pred Value : 0.8913
Neg Pred Value : 0.8740
Prevalence : 0.5555
Detection Rate : 0.5002
Detection Prevalence : 0.5612
Balanced Accuracy : 0.8816
'Positive' Class : TRUE
We can plot the predicted outcome spatially by extracting the sdf output from the model as an sf object.
gwr.fixed.sf <- st_as_sf(gwlr.fixed$SDF)estprob <- tm_shape(osun)+
tm_polygons(alpha=0.1)+
tm_text(text="ADM2_EN")+
tm_shape(gwr.fixed.sf) +
tm_dots(col="yhat",
border.col = "gray60",
border.lwd = 1,
palette = "YlOrRd") +
tm_view(set.zoom.limits = c(9, 12))
actual <- tm_shape(osun)+
tm_polygons(alpha=0.1)+
tm_text(text="ADM2_EN")+
tm_shape(gwr.fixed.sf) +
tm_dots(col="y",
border.col = "gray60",
border.lwd = 1,
palette = c("#FFFFB2", "#BD0026")) +
tm_view(set.zoom.limits = c(9, 12))
tmap_arrange(actual, estprob,
asp=1, ncol=2,
sync = TRUE)Refining the Model
We can further refine the model by removing the variables (distance_to_primary_road, distance_to_secondary_road ) that we previously identified as not statistically significant. To do that, we create a new formula without those variables. We then need to find the recommended fixed bandwidth for the formula.
fm2 <- as.formula(paste("status ~",
paste(expvars[4:11], collapse="+")))
bw.fixed <- bw.ggwr(fm2,
data=wp_clean_sp,
family = "binomial",
approach= "AIC",
kernel = "gaussian",
adaptive = FALSE,
longlat= FALSE)Take a cup of tea and have a break, it will take a few minutes.
-----A kind suggestion from GWmodel development group
Iteration Log-Likelihood:(With bandwidth: 95768.67 )
=========================
0 -2890
1 -2837
2 -2830
3 -2829
4 -2829
5 -2829
Fixed bandwidth: 95768.67 AICc value: 5681.18
Iteration Log-Likelihood:(With bandwidth: 59200.13 )
=========================
0 -2878
1 -2820
2 -2812
3 -2810
4 -2810
5 -2810
Fixed bandwidth: 59200.13 AICc value: 5645.901
Iteration Log-Likelihood:(With bandwidth: 36599.53 )
=========================
0 -2854
1 -2790
2 -2777
3 -2774
4 -2774
5 -2774
6 -2774
Fixed bandwidth: 36599.53 AICc value: 5585.354
Iteration Log-Likelihood:(With bandwidth: 22631.59 )
=========================
0 -2810
1 -2732
2 -2711
3 -2707
4 -2707
5 -2707
6 -2707
Fixed bandwidth: 22631.59 AICc value: 5481.877
Iteration Log-Likelihood:(With bandwidth: 13998.93 )
=========================
0 -2732
1 -2635
2 -2604
3 -2597
4 -2596
5 -2596
6 -2596
Fixed bandwidth: 13998.93 AICc value: 5333.718
Iteration Log-Likelihood:(With bandwidth: 8663.649 )
=========================
0 -2624
1 -2502
2 -2459
3 -2447
4 -2446
5 -2446
6 -2446
7 -2446
Fixed bandwidth: 8663.649 AICc value: 5178.493
Iteration Log-Likelihood:(With bandwidth: 5366.266 )
=========================
0 -2478
1 -2319
2 -2250
3 -2225
4 -2219
5 -2219
6 -2220
7 -2220
8 -2220
9 -2220
Fixed bandwidth: 5366.266 AICc value: 5022.016
Iteration Log-Likelihood:(With bandwidth: 3328.371 )
=========================
0 -2222
1 -2002
2 -1894
3 -1838
4 -1818
5 -1814
6 -1814
Fixed bandwidth: 3328.371 AICc value: 4827.587
Iteration Log-Likelihood:(With bandwidth: 2068.882 )
=========================
0 -1837
1 -1528
2 -1357
3 -1261
4 -1222
5 -1222
Fixed bandwidth: 2068.882 AICc value: 4772.046
Iteration Log-Likelihood:(With bandwidth: 1290.476 )
=========================
0 -1403
1 -1016
2 -807.3
3 -680.2
4 -680.2
Fixed bandwidth: 1290.476 AICc value: 5809.721
Iteration Log-Likelihood:(With bandwidth: 2549.964 )
=========================
0 -2019
1 -1753
2 -1614
3 -1538
4 -1506
5 -1506
Fixed bandwidth: 2549.964 AICc value: 4764.056
Iteration Log-Likelihood:(With bandwidth: 2847.289 )
=========================
0 -2108
1 -1862
2 -1736
3 -1670
4 -1644
5 -1644
Fixed bandwidth: 2847.289 AICc value: 4791.834
Iteration Log-Likelihood:(With bandwidth: 2366.207 )
=========================
0 -1955
1 -1675
2 -1525
3 -1441
4 -1407
5 -1407
Fixed bandwidth: 2366.207 AICc value: 4755.524
Iteration Log-Likelihood:(With bandwidth: 2252.639 )
=========================
0 -1913
1 -1623
2 -1465
3 -1376
4 -1341
5 -1341
Fixed bandwidth: 2252.639 AICc value: 4759.188
Iteration Log-Likelihood:(With bandwidth: 2436.396 )
=========================
0 -1980
1 -1706
2 -1560
3 -1479
4 -1446
5 -1446
Fixed bandwidth: 2436.396 AICc value: 4756.675
Iteration Log-Likelihood:(With bandwidth: 2322.828 )
=========================
0 -1940
1 -1656
2 -1503
3 -1417
4 -1382
5 -1382
Fixed bandwidth: 2322.828 AICc value: 4756.471
Iteration Log-Likelihood:(With bandwidth: 2393.017 )
=========================
0 -1965
1 -1687
2 -1539
3 -1456
4 -1422
5 -1422
Fixed bandwidth: 2393.017 AICc value: 4755.57
Iteration Log-Likelihood:(With bandwidth: 2349.638 )
=========================
0 -1949
1 -1668
2 -1517
3 -1432
4 -1398
5 -1398
Fixed bandwidth: 2349.638 AICc value: 4755.753
Iteration Log-Likelihood:(With bandwidth: 2376.448 )
=========================
0 -1959
1 -1680
2 -1530
3 -1447
4 -1413
5 -1413
Fixed bandwidth: 2376.448 AICc value: 4755.48
Iteration Log-Likelihood:(With bandwidth: 2382.777 )
=========================
0 -1961
1 -1683
2 -1534
3 -1450
4 -1416
5 -1416
Fixed bandwidth: 2382.777 AICc value: 4755.491
Iteration Log-Likelihood:(With bandwidth: 2372.536 )
=========================
0 -1958
1 -1678
2 -1528
3 -1445
4 -1411
5 -1411
Fixed bandwidth: 2372.536 AICc value: 4755.488
Iteration Log-Likelihood:(With bandwidth: 2378.865 )
=========================
0 -1960
1 -1681
2 -1532
3 -1448
4 -1414
5 -1414
Fixed bandwidth: 2378.865 AICc value: 4755.481
Iteration Log-Likelihood:(With bandwidth: 2374.954 )
=========================
0 -1959
1 -1679
2 -1530
3 -1446
4 -1412
5 -1412
Fixed bandwidth: 2374.954 AICc value: 4755.482
Iteration Log-Likelihood:(With bandwidth: 2377.371 )
=========================
0 -1959
1 -1680
2 -1531
3 -1447
4 -1413
5 -1413
Fixed bandwidth: 2377.371 AICc value: 4755.48
Iteration Log-Likelihood:(With bandwidth: 2377.942 )
=========================
0 -1960
1 -1680
2 -1531
3 -1448
4 -1414
5 -1414
Fixed bandwidth: 2377.942 AICc value: 4755.48
Iteration Log-Likelihood:(With bandwidth: 2377.018 )
=========================
0 -1959
1 -1680
2 -1531
3 -1447
4 -1413
5 -1413
Fixed bandwidth: 2377.018 AICc value: 4755.48 bw.fixed[1] 2377.371gwlr2.fixed <- ggwr.basic(fm2,
data=wp_clean_sp,
bw = 2377.371,
family = "binomial",
kernel = "gaussian",
adaptive = FALSE,
longlat= FALSE) Iteration Log-Likelihood
=========================
0 -1959
1 -1680
2 -1531
3 -1447
4 -1413
5 -1413 gwlr2.fixed ***********************************************************************
* Package GWmodel *
***********************************************************************
Program starts at: 2022-12-18 13:46:51
Call:
ggwr.basic(formula = fm2, data = wp_clean_sp, bw = 2377.371,
family = "binomial", kernel = "gaussian", adaptive = FALSE,
longlat = FALSE)
Dependent (y) variable: status
Independent variables: distance_to_tertiary_road distance_to_city distance_to_town water_point_population local_population_1km usage_capacity is_urban water_source_clean
Number of data points: 4756
Used family: binomial
***********************************************************************
* Results of Generalized linear Regression *
***********************************************************************
Call:
NULL
Deviance Residuals:
Min 1Q Median 3Q Max
-129.368 -1.750 1.074 1.742 34.126
Coefficients:
Estimate Std. Error z value Pr(>|z|)
Intercept 3.540e-01 1.055e-01 3.354 0.000796
distance_to_tertiary_road 1.001e-04 2.040e-05 4.910 9.13e-07
distance_to_city -1.764e-05 3.391e-06 -5.202 1.97e-07
distance_to_town -1.544e-05 2.825e-06 -5.466 4.60e-08
water_point_population -5.098e-04 4.476e-05 -11.390 < 2e-16
local_population_1km 3.452e-04 1.779e-05 19.407 < 2e-16
usage_capacity1000 -6.206e-01 6.966e-02 -8.908 < 2e-16
is_urbanTRUE -2.667e-01 7.474e-02 -3.569 0.000358
water_source_cleanProtected Shallow Well 4.947e-01 8.496e-02 5.823 5.79e-09
water_source_cleanProtected Spring 1.279e+00 4.384e-01 2.917 0.003530
Intercept ***
distance_to_tertiary_road ***
distance_to_city ***
distance_to_town ***
water_point_population ***
local_population_1km ***
usage_capacity1000 ***
is_urbanTRUE ***
water_source_cleanProtected Shallow Well ***
water_source_cleanProtected Spring **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 6534.5 on 4755 degrees of freedom
Residual deviance: 5688.9 on 4746 degrees of freedom
AIC: 5708.9
Number of Fisher Scoring iterations: 5
AICc: 5708.923
Pseudo R-square value: 0.129406
***********************************************************************
* Results of Geographically Weighted Regression *
***********************************************************************
*********************Model calibration information*********************
Kernel function: gaussian
Fixed bandwidth: 2377.371
Regression points: the same locations as observations are used.
Distance metric: A distance matrix is specified for this model calibration.
************Summary of Generalized GWR coefficient estimates:**********
Min. 1st Qu. Median
Intercept -3.7021e+02 -4.3797e+00 3.5590e+00
distance_to_tertiary_road -3.1622e-02 -4.5462e-04 9.1291e-05
distance_to_city -5.4555e-02 -6.5623e-04 -1.3507e-04
distance_to_town -8.6549e-03 -5.2754e-04 -1.6785e-04
water_point_population -2.9696e-02 -2.2705e-03 -1.2277e-03
local_population_1km -7.7730e-02 4.4281e-04 1.0548e-03
usage_capacity1000 -5.5889e+01 -1.0347e+00 -4.1960e-01
is_urbanTRUE -7.3554e+02 -3.4675e+00 -1.6596e+00
water_source_cleanProtected.Shallow.Well -1.8842e+02 -4.7295e-01 6.2378e-01
water_source_cleanProtected.Spring -1.3630e+03 -5.3436e+00 2.7714e+00
3rd Qu. Max.
Intercept 1.3755e+01 2171.6375
distance_to_tertiary_road 6.3011e-04 0.0237
distance_to_city 1.5921e-04 0.0162
distance_to_town 2.4490e-04 0.0179
water_point_population 4.5879e-04 0.0765
local_population_1km 1.8479e-03 0.0333
usage_capacity1000 3.9113e-01 9.2449
is_urbanTRUE 1.0554e+00 995.1841
water_source_cleanProtected.Shallow.Well 1.9564e+00 66.8914
water_source_cleanProtected.Spring 7.0805e+00 208.3749
************************Diagnostic information*************************
Number of data points: 4756
GW Deviance: 2815.659
AIC : 4418.776
AICc : 4744.213
Pseudo R-square value: 0.5691072
***********************************************************************
Program stops at: 2022-12-18 13:49:55 We should also compare the confusion matrix of the new local model with the first local model.
gwr2.fixed <- as.data.frame(gwlr2.fixed$SDF) %>%
mutate(most = as.factor(
ifelse(
yhat >= 0.5, T, F)),
y = as.factor(y)
)
CMConfusion Matrix and Statistics
Reference
Prediction FALSE TRUE
FALSE 1824 263
TRUE 290 2379
Accuracy : 0.8837
95% CI : (0.8743, 0.8927)
No Information Rate : 0.5555
P-Value [Acc > NIR] : <2e-16
Kappa : 0.7642
Mcnemar's Test P-Value : 0.2689
Sensitivity : 0.9005
Specificity : 0.8628
Pos Pred Value : 0.8913
Neg Pred Value : 0.8740
Prevalence : 0.5555
Detection Rate : 0.5002
Detection Prevalence : 0.5612
Balanced Accuracy : 0.8816
'Positive' Class : TRUE
CM2 <- confusionMatrix(data=gwr2.fixed$most,
positive = "TRUE",
reference = gwr2.fixed$y)
CM2Confusion Matrix and Statistics
Reference
Prediction FALSE TRUE
FALSE 1833 268
TRUE 281 2374
Accuracy : 0.8846
95% CI : (0.8751, 0.8935)
No Information Rate : 0.5555
P-Value [Acc > NIR] : <2e-16
Kappa : 0.7661
Mcnemar's Test P-Value : 0.6085
Sensitivity : 0.8986
Specificity : 0.8671
Pos Pred Value : 0.8942
Neg Pred Value : 0.8724
Prevalence : 0.5555
Detection Rate : 0.4992
Detection Prevalence : 0.5582
Balanced Accuracy : 0.8828
'Positive' Class : TRUE
gwr2.fixed <- gwr2.fixed %>%
mutate(most2 = as.factor(
ifelse(
yhat >= 0.6, T, F))
)
CM3 <- confusionMatrix(data=gwr2.fixed$most2,
positive = "TRUE",
reference = gwr2.fixed$y)
CM3Confusion Matrix and Statistics
Reference
Prediction FALSE TRUE
FALSE 1976 472
TRUE 138 2170
Accuracy : 0.8717
95% CI : (0.8619, 0.8811)
No Information Rate : 0.5555
P-Value [Acc > NIR] : < 2.2e-16
Kappa : 0.7443
Mcnemar's Test P-Value : < 2.2e-16
Sensitivity : 0.8213
Specificity : 0.9347
Pos Pred Value : 0.9402
Neg Pred Value : 0.8072
Prevalence : 0.5555
Detection Rate : 0.4563
Detection Prevalence : 0.4853
Balanced Accuracy : 0.8780
'Positive' Class : TRUE
Now, let’s plot the predicted results of the second model spatially.
gwr2.fixed.sf <- st_as_sf(gwlr2.fixed$SDF) estprob <- tm_shape(osun)+
tm_polygons(alpha=0.1)+
tm_text(text="ADM2_EN")+
tm_shape(gwr2.fixed.sf) +
tm_dots(col="yhat",
border.col = "gray60",
border.lwd = 1,
palette = "YlOrRd") +
tm_view(set.zoom.limits = c(9, 12))
tmap_arrange(actual, estprob,
asp=1, ncol=2,
sync = TRUE)We should also directly compare the prediction result with the actual. The code chunk below adds the prediction results based on the 0.6 threshold level. We also add indicators for false negatives and false positives to see if the misclassifications show spatial depedency.
gwr2.fixed.sf <- gwr2.fixed.sf%>%
mutate(thres0.6 = as.factor(
ifelse(yhat >= 0.6, T, F)),
y = as.factor(y),
FP = ifelse(thres0.6==T & y==F, T, F),
FN = ifelse(thres0.6==F & y==T, T, F)
)pred0.6 <- tm_shape(osun)+
tm_polygons(alpha=0.1)+
tm_text(text="ADM2_EN")+
tm_shape(gwr2.fixed.sf) +
tm_dots(col="thres0.6",
border.col = "gray60",
border.lwd = 1,
palette = c("#FFFFB2", "#BD0026")) +
tm_view(set.zoom.limits = c(9, 12))
FN <- tm_shape(osun)+
tm_polygons(alpha=0.1)+
tm_text(text="ADM2_EN")+
tm_shape(filter(gwr2.fixed.sf, FN==T)) +
tm_dots(col="FN",
border.col = "gray60",
border.lwd = 1,
palette = c("#FFFFFF", "#000000")) +
tm_view(set.zoom.limits = c(9, 12))
FP <- tm_shape(osun)+
tm_polygons(alpha=0.1)+
tm_text(text="ADM2_EN")+
tm_shape(filter(gwr2.fixed.sf, FP==T)) +
tm_dots(col="FP",
border.col = "gray60",
border.lwd = 1,
palette = c("#FFFFFF", "#000000")) +
tm_view(set.zoom.limits = c(9, 12))
tmap_arrange(actual, pred0.6, FP, FN,
asp=1, ncol=2,
sync = TRUE)